David S. Tourigny
Birmingham Fellow
School of Mathematics, University of Birmingham
I am a Birmingham Fellow (Assistant Professor equivalent) within the School of Mathematics, University of Birmingham. Prior to joining Birmingham, I spent five years as a Postdoctoral Fellow of the Simons Foundation and then Associate Research Scientist at Columbia University (Irving Medical Centre and Irving Institute for Cancer Dynamics). Before that, I was a Research Fellow (Peterhouse) in the Department of Applied Mathematics and Theoretical Physics, University of Cambridge where I also completed my PhD (Trinity College) under the supervision of Venki Ramakrishnan and Garib Murshudov (MRC Laboratory of Molecular Biology).
Research
Publications
Here is a short list of selected publications. A full list can be found via Google Scholar
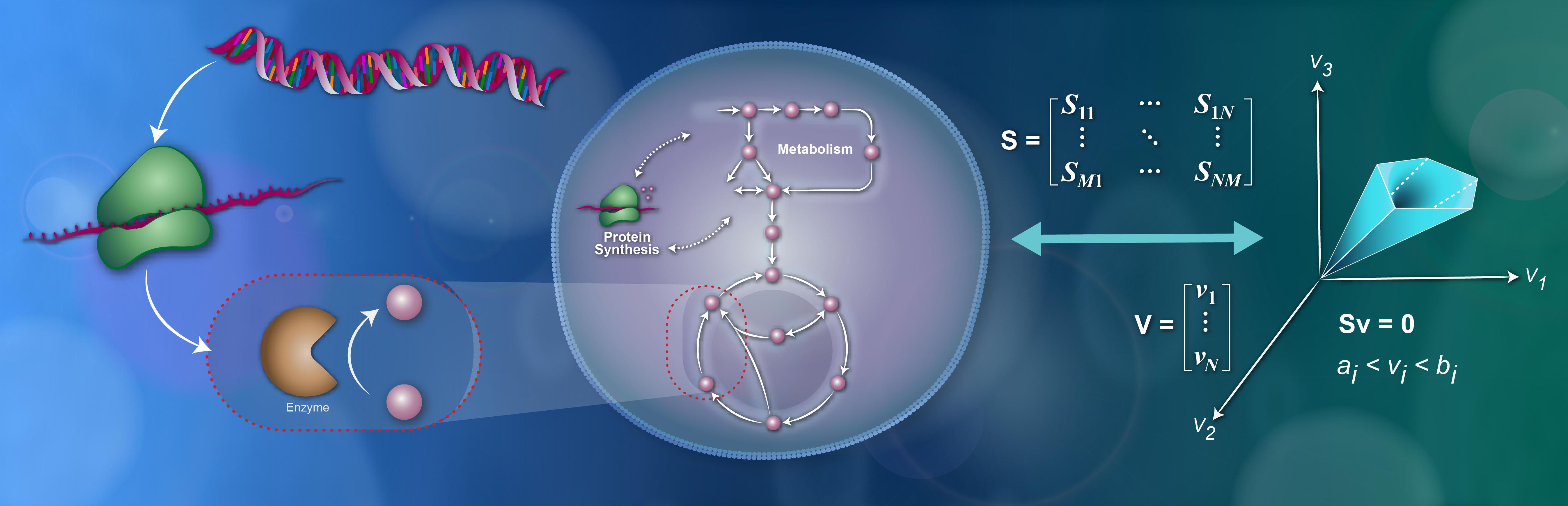
- Metabolic coordination and phase transitions in spatially distributed multi-cellular systems
- Cellular landscape of adrenocortical carcinoma at single-nuclei resolution
- Molecular characterization of the tumor microenvironment in renal medullary carcinoma
- Simulating single-cell metabolism using a stochastic flux-balance analysis algorithm
- Cooperative metabolic resource allocation in spatially-structured systems
- Dynamic metabolic resource allocation based on the maximum entropy principle
I have also contributed three chapters to the text book Economic Principles in Cell Biology.
Software
I develop and maintain the Python/C++ package dfba (see also paper here) for dynamic flux-balance analysis (DFBA) simulations. This is joint work with Moritz E. Beber and Jorge Carrasco Muriel that forms part of the openCOBRA code base for constraint-based reconstruction and analysis of metabolic models.
Other software and scripts for data processing can be accessed via GitLab.
During my PhD I worked on a maximum likelihood-based algorithm for clustering and inferring diffraction data from macromolecular crystals (today, this would probably be called machine learning). Details can be found in the second section of my thesis.
Teaching and supervision
Current and former research students:
- Linus Chang (PhD Applied Mathematics, current)
- Thomas Munn (MSci Mathematics, current)
- Balqees Shaaban (MSc Bioinformatics)
- Kirsty Richens (MSci Mathematics)
- Thomas Cong (STS winner)
- LM Advanced Mathematical Biology (31128) at University of Birmingham. Lecture notes here
- Mathematics group study lead for REUK
- Lectures on Mathematical Foundations of Constraint-Based Modelling
Teaching: